Cell’s Design Helps Launch Quick Defenses When under Attack
Immune system cells found to reposition key genes to rapidly fight invading pathogens
February 11, 2020
By Mario Aguilera
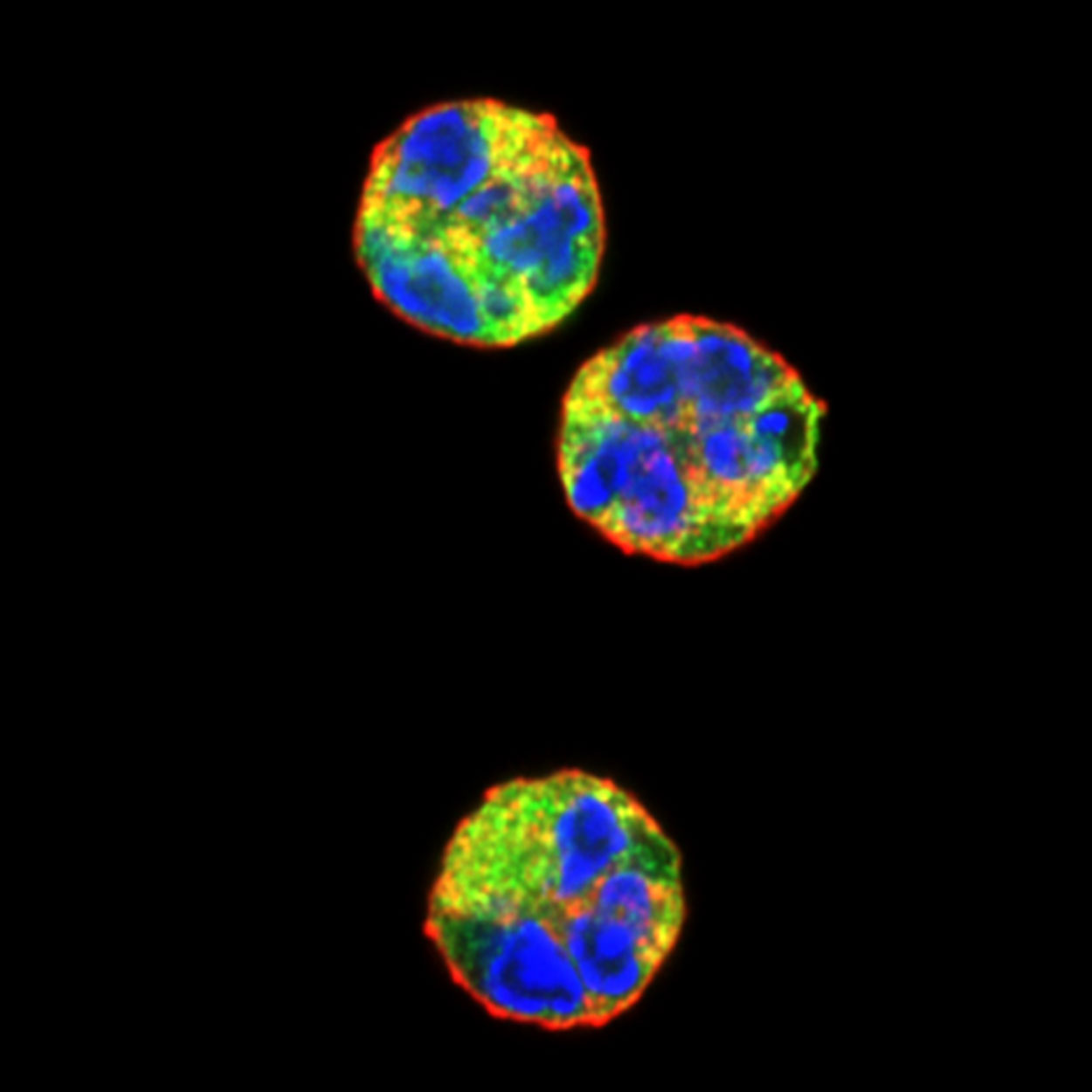
Confocal fluorescent microscopy images of unstimulated human neutrophils stained with wheat germ agglutinin to label the plasma membrane (red), anti-neutrophil aquaporin AQP9 antibodies to label the cytoplasm (green) and DAPI to label DNA (blue). The colors highlight the unique morphology of the neutrophil nucleus.
Under normal, healthy conditions, genes within immune system cells known as neutrophils remain dormant.
But once an infection is detected, neutrophils sound the alarm and push these genes into action as a critical line of defense against invading pathogens. These genes help the cell fight back by releasing an array of molecules that attack infection-causing bacteria.
Although neutrophils were discovered more than a century ago, until now the measures underlying their immune defenses, or “inflammatory response,” has remained a mystery.
In a study recently published in Genes and Development, scientists at the University of California San Diego now have a clearer picture of what’s involved in this process.
Researchers led by Distinguished Professor Cornelis Murre’s laboratory found that the design and architecture of the cell’s nucleus plays a key role in helping neutrophils spring into defensive action. Prior to a threat, the genes within neutrophils that are needed for a defensive response are purposely kept well away in the nuclear envelope area, a design that prevents unnecessary activation and inflammation.
But once neutrophils detect a bacterial invasion, they quickly reposition these genes to the interior of the nucleus, where they can become expressed as an inflammatory response in a matter of hours. The key is a folding process that repositions defensive genes from the cell outlands to the center of action.
“It’s a very elegant mechanism that’s tightly controlled to make sure these genes are being properly regulated,” said Murre, a researcher in the Division of Biological Sciences’ Section of Molecular Biology. “This explains how neutrophils remain dormant in the absence of bacteria, but they are positioned so they readily move after sensing bacteria in the neighborhood.”
Murre says the same processes may be involved in tumors, which could be an area of future investigations.
The architecture and folding phenomenon found in the current study has been seen before. Stem cells that change to become B or T cells similarly go through a repositioning of genes during development from the nuclear envelope to the interior.
“These are similar mechanisms,” said Murre. “Nature always uses the same thing over and over again. Now we see that genes move not only to induce gene expression during development but we see them move during an inflammatory response.”
Coauthors of the paper are: Matthew Denholtz, Yina Zhu, Zhaoren He (graduate student), Hanbin Lu (graduate student), Takeshi Isoda, Simon Döhrmann, Victor Nizet and Cornelis Murre.
Funding for the study was provided by the Center for Computational Biology and Bioinformatics (UL1TRR001442); the California Institute for Regenerative Medicine (RB5-07025); the National Institutes of Health (AI082850, AI00880, AI09599 and 1 U01 AI124316); the Frontiers of Innovation Scholars Program; the Waitt Advanced Biophotonics Core Facility of the Salk Institute with funding from the National Institutes of Health-National Cancer Institute Cancer Center Support Grant (P30 014195); the Waitt Foundation; and the Uehara Memorial Foundation.