1. TIGR has reannotated the MIPS data, and in the process has redefined many of the genes. Therefore, MIPS AGI names do not not always refer to the same gene as TIGR AGI names. (Be aware of this when using data from other sources; many researchers are still using the MIPS identifiers.)
2. Affymetrix has released the target sequences for the probe sets on the 8K Genechip. Because these sequences were not available to us at the time our original files were created, our original AGI assignments were based on the gene identifiers in the descriptions provided by Affymetrix. Comparing the target sequences with the TIGR genome data reveals that many of the original gene assignments from Affymetrix do not correspond to the best sequence match in TIGR.
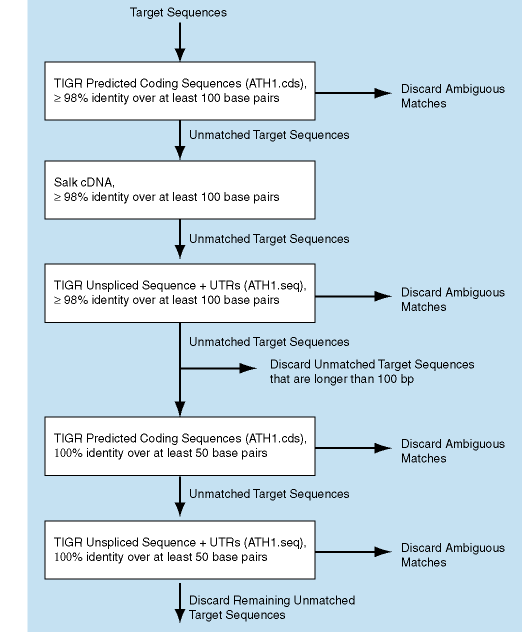
AGI assignments for the probe sets were obtained by BLASTing the Affymetrix target sequences against the following data sources:
At each step probe sets with ambiguous matches were discarded.

Using these criteria, we were able to identify unambiguous matches for 95% of the probe sets. At each step we have attempted to be very conservative. If there was any doubt about which gene was the best match for a given probe set, we did not assign an AGI identifier. For each probe set, our annotation file describes the source and criterion for that AGI assignment.
Updated: 8/1/2002